Abstract
Objective: analysis of primary drug resistance of HIV in adult patients from the Republic of Guinea
Design: Cross-sectional study.
Methods: 2,168 residents of the Republic of Guinea (blood donors, as well as Rusal employees and their families who were undergoing routine medical examination) were tested for the presence of HIV serological markers using ELISA. Individuals with a positive result were further examined for the presence of viral load (VL) in blood plasma.
If VL was high enough (> 500 copies / ml), HIV was tested using Sanger sequencing. The obtained sequences were genotyped using the REGA HIV-1 Subtyping Tool - Version 3.0 and phylogenetic analysis in the MEGA 10 program. Analysis for the presence of mutations associated with drug resistance was carried out using the Stanford University HIV Drug Resistance Database.
Results: Serological markers of HIV were detected in 239 people, which was 11.02% of the complete sample. HIV RNA was detected in 58 people.
Genotyping revealed the prevalence of circulating recombinant HIV CRF02_AG (41.9%) in the study group, as well as other subtypes: A1 (29.1%); A3 (12.9%); non-CRF02_AG recombinant between A1 and G (12.9%); subtype G (3.2%).
In 25% of patients, at least one significant mutation was encountered, leading directly to drug resistance of HIV for their virus subtype. The mutations encountered cause resistance to nucleoside and non-nucleoside reverse transcriptase inhibitors; one case of multiple resistance was identified. Major resistance to IP was not met.
Conclusions: Thus, the genetic structure of HIV in the Republic of Guinea is close to that described in the literature for West African countries. The incidence of mutations associated with HIV resistance to ARVs was relatively high. Given the increasing number of patients who start taking ART, the high incidence of primary drug resistance can lead to frequent cases of therapy failure and, as a result, change of treatment regimens. To prescribe effective treatment regimens, it is necessary to introduce studies on the presence of primary resistance in newly diagnosed patients.
Key words: HIV-1 transmitted drug resistance, West Africa, Guinea Republic
Introduction
The human immunodeficiency virus (HIV) epidemic continues to spread rapidly around the world. According to The Joint United Nations Programme on HIV/AIDS (UNAIDS) estimates, currently the number of people infected with HIV on the Earth is about 31.6 - 44.5 million people, while the number of new infections amounted to 1.2 - 2.2 million cases in 2019 [1]. The African continent is one of the most HIV-affected territories in the world, currently home to 25.6 million people living with HIV (PLHIV), which is 67.37% of all registered HIV-infected in the world. The overwhelming majority (80.86%) are in the countries of Eastern and Southern Africa [2].
The Republic of Guinea is a located in West Africa. The HIV prevalence in Guinea was approximately 1.6% in 2014 and has practically unchanged by 2019, but treatment coverage also remains one of the lowest in the world, with less than a quarter (23%) of people living with HIV on antiretroviral therapy (ART) [1, 3].
Among other things, Africa, especially Western and Central, has the highest diversity of HIV subtypes in the world. In West and Central Africa, the most common circulating recombinant form (CRF) 02_AG. The A and G subtypes co-circulate in some countries such as Nigeria [4, 5, 6]. The study in 2016 confirmed the prevalence of CRF 02_AG in Guinea [7].
The Republic of Guinea is one of the countries affected by the Ebola epidemic, and therefore in the period 2014-2015, there were significant difficulties in providing treatment to patients with HIV infection. This, in turn, could affect the prevalence of drug resistance (DR), as a result of which more careful monitoring of HIV resistance is required in this region. However, the most recent data on drug resistance in Guinea reported in the literature dates back to 2009, when the prevalence of primary drug resistance was 8.9 % [ссылка]. In 2016, it was not possible to analyze a sufficient number of samples to identify the prevalence level of DR in the Republic of Guinea [7].
Methods
Study setting
During the study, 2,168 blood plasma samples were obtained from healthy people from the Republic of Guinea: blood donors and employees of UC RUSAL and their families with no data about disease caused by the Ebola virus. The surveyed persons denied a history of HIV infection. The study was conducted based of the Center of Epidemiology and Microbiology (CREMS) in Kindia. The material was collected by UC RUSAL employees and Institute of Applied Biological Research of Guinea (IRBAG) specialists.
Participant Enrollment
Inclusion criteria were: (i) ≥18 years (ii) no history of HIV infection
Laboratory Methods
Patients were examined for the presence of HIV antigens and antibodies to the virus by ELISA. Quantitative analysis of HIV RNA was carried out with a commercial kit "AmpliSens® HIV-Monitor-FRT" (Central Research Institute of Epidemiology, Russia) with a sensitivity threshold of 500 copies/ml, and samples with detectable viral load (VL) were analyzed using RT-PCR and Sanger sequencing.
For reverse transcription and amplification of HIV, commercial kits "RT-PCR-kit-Pro/Rev" and "PCR-kit-Pro/Rev" (Central Research Institute of Epidemiology, Russia) were used, the sequencing reaction was performed according to the instructions for the kit "AmpliSens® HIVResist-Seq" (Central Research Institute of Epidemiology, Russia) [как было показано ранее или типа того и ссылка на что-то из наших статей].
For HIV genotyping we used nucleotide sequences of 1302 nt spanning the pol gene in the range of 2253–3554 nt., Coordinates are given for the data presented in the international GenBank database HIV HXB2 (K03455.1). The analysis of the sequencing reaction products was performed using an ABI Prism 3500 genetic analyzer (Applied Biosystems, USA).
Nucleotide sequences were analyzed using the NCBI Blast program in comparison with nucleotide sequences presented in the international GenBank database. Alignment of nucleotide sequences was executed with MEGAv.7.0 program using the ClustalW algorithm [8]. For the construction of phylogenetic trees and subsequent phylogenetic analysis, the Neighbor-joining algorithm was used, which allows optimization of trees by the criterion of “balanced minimum evolution”; when assessing the reliability of phylogenetic relationships, we used multiple generations of samples using the Bootstrap method for 1000 independent constructions of each phylogenetic tree.
Isolates were genotyped using the REGA HIV-1 Subtyping Tool 3.0 program and analysis of their phylogenetic relationships with reference sequences from the GenBank international database (Fig. 1), which made it possible to more accurately assess the distribution of HIV-1 subtypes in the studied population. Analysis of HIV genetic sequences for the presence of drug resistance mutations was performed using the Stanford HIV Data Base
Results
Study Population
Суммарно был исследован материл от 2168 пациентов, возраст которых находился в пределах 18-58 лет, медианный возраст составил 38 лет (β = 0.95, (37.58; 39.72)). Большинство обследованных пациентов – мужского пола (68.17% (β = 0.95; (66.21%; 70.13%))).
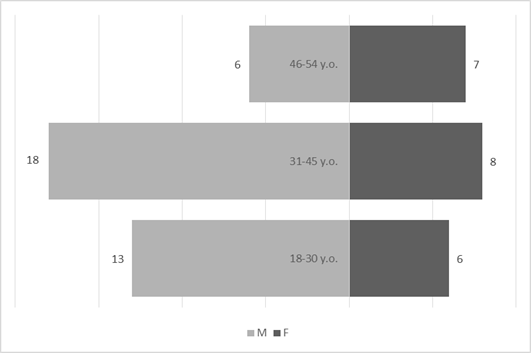
Genotyped samples were obtained from patients aged 18 to 54 years. The median age was 36 years (β = 0.95, (32.53; 39.47)). The predominant age group is from 31 to 45 years old (62.07%). The most represented in the sample are men, they make up 63.79% of the study group (β = 0.95, (52%; 76%)) compared with women - 36.21% (β = 0.95, (24%; 49%)).
Discussion
There is a significant discrepancy between the number of HIV-positive people identified using ELISA and those who are found to have viral RNA. Moreover, the percentage of people in whom the virus was detected by the PCR method correlates with the literature data on the prevalence of HIV infection in the Republic of Guinea. However, the significant percentage of apparently healthy people who are HIV seropositive suggests that the incidence of HIV may be higher than the literature suggests. However, to confirm this, additional screening studies with a confirmatory test are necessary.
Выявленное генетическое разнообразие соотносится с литературными сведениями о субсубтипах ВИЧ, циркулирующих в Западной Африке и высоким генетическим разнообразием ВИЧ в Африке в целом. В подавляющем большинстве случаев встречается циркулирующая рекомбинантная форма 02_AG. При этом особое внимание необходимо уделить изолятам, генотипирование которых показывает не столь однозначные результаты. Для создания более полной картины мы дополнительно провели филогенетический анализ совместно с изолятами, полученными в исследовании 2016 года. На обоих филогенетических древах выделился образец 18, субтипированный в REGA 3.0 как рекомбинант генотипов A1 и G, но не принадлежащий к CRF02_AG. Интересно отметить, что он образует кластер с изолятом, депонированным под номером LT976766, который в работе 2016 года отнесен к CRF02_AG, что можно объяснить нехваткой информации о последовательности гена протеазы у данного образца. Важно отметить, что образцы 44 и 46, определенных в REGA 3.0 как неизвестные рекомбинантные формы, в обоих филогенетических древах наиболее близко кластеризуются с CRF02_AG. Следует отметить, что образцы 44, 46 и 13, согласно результатам анализа в REGA HIV-1, также являются рекомбинантами между известным CRF_02AG и подтипом A1 (рис. 4).
Исходя из результатов проведенного анализа, мы предполагаем значительный вклад различных рекомбинантных форм ВИЧ в генетическое разнообразие вируса в исследуемом регионе. В связи с этим крайне важным является изучение полных геномов ВИЧ в Гвинейской республике для выявления всех имеющихся в данном регионе рекомбинантных форм и наиболее встречающихся точек рекомбинации.
Genetic diversity of HIV in the surveyed group correlates with what is usually observed in West African countries - in the vast majority of cases, CRF_02AG is found. However, other recombinant forms circulate simultaneously with it. Samples 18, 44, and 46 deserve special attention; they separated from the main branch CRF_02AG on the phylogenetic tree. Analysis of their nucleotide sequence using the REGA HIV-1 subtyping tool showed that these samples are recombinants of genotypes A1 and G, but do not belong to CRF02_AG (Fig 3). Interestingly, sample 13 is also a recombinant of genotypes A1 and G but does not belong to CRF02_AG, although it is located on the CRF02_AG branch. It should be noted that samples 44, 46, and 13, according to the results of the analysis in REGA HIV-1, are also recombinants between the known CRF_02AG and subtype A1 (fig 4).
For some samples, subtyping is difficult. Since in the framework of this study only the region of the pol gene was studied, it is impossible to explore the points of recombination outside the studied fragment. However, the discrepancy between the results of genotyping using the REGA 3.0 tool and our own phylogenetic analysis raises suspicions of a greater part of recombinant forms of the virus to the genetic diversity of HIV in the Republic of Guinea.
Since accurate genotyping is critical to screening a sample for resistant HIV variants, more research is needed to assess the contribution of recombinant forms to the genetic diversity of the virus in the region. Insufficient attention to the high diversity of HIV recombinants and the lack of complete data on common points of recombination can lead to an erroneous determination of the presence or absence of the virus DR.
The incidence of mutations associated with HIV resistance to ARVs was relatively high. Given the increasing number of patients who start taking ART, the high incidence of primary drug resistance can lead to frequent cases of therapy failure and, as a result, change of treatment regimens. To prescribe effective treatment regimens, it is necessary to introduce studies on the presence of primary resistance in newly diagnosed patients.
A similar high incidence of drug resistance mutations in untreated infected individuals has been shown in Sierra Leone, which is associated with disruptions to HIV services during the Ebola epidemic in 2014-2016 since the study was conducted in a close geographic region (Liberia) before the epidemic showed a significantly lower incidence of such cases [10].
Выявленные у наивных пациентов 24 мутации на более обширной выборке, скорее всего, имели бы приблизительно равные частоты встречаемости, что ожидаемо, так как в исследуемой группе отсутствует давление на вирус антиретровирусными препаратами, и мутации спонтанно возникают и исчезают в поколениях вируса.
It is interesting to note that in more than half of the cases (72 %) there were substitutions in the twentieth position. Можно заметить, что из этих мутаций наиболее выделяется K20I - is a polymorphic variant for subtypes CRF_02AG and G, but reduces sensitivity to nelfinavir in subtypes B and C. However, there is evidence that this mutation may enhance viral replication in non-B subtypes [11]. Данная мутация имеет встречаемость значительно более высокую, чем все остальные (67.24 % (β = 0.95, (53,66%; 78,99%)) несмотря на отсутствие факторов отбора в виде АРВТ, что может быть следствием преимущества, получаемого носителями данной мутации.
A non-polymorphic variant of K20V has also been identified, which is rare, and its effect on HIV susceptibility to ARVs is poorly understood. Also, two mutations in the tenth position were identified, one of which, detected in one case, L10LF, is a minor mutation of resistance to PI, and the other, found in 29.03% of cases, L10I, increases the replication of viruses with other resistance mutations to SP [12]. Two rare mutations were identified: M46M_M and N88NH, in the positions of which resistance mutations to protease inhibitors are found, however, these variants have not been described among them.
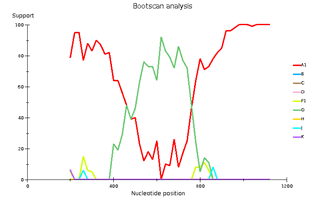
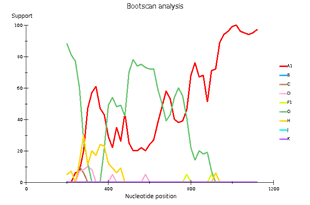
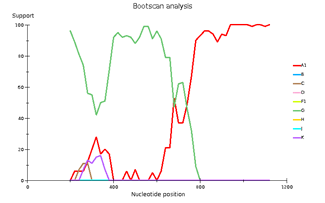
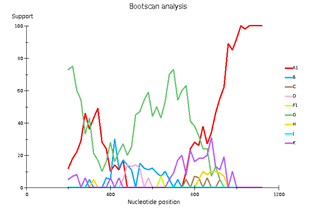
Fig 3. Subtype recombination analysis with REGA HIV-1 Subtyping Tool 3.0. Bootscan analysis performed with window size 400 and step size 20.
A – sample 18; B – sample 44; C – sample 46; D – sample 13.
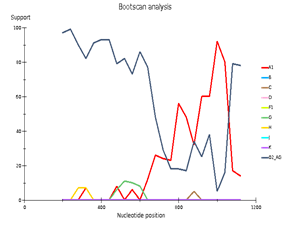
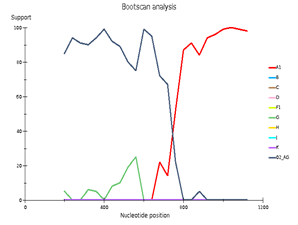
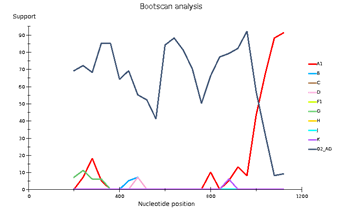
Fig 4. CRF/Subtype Recombination Analysis with REGA HIV-1 Subtyping Tool 3.0. Bootscan analysis performed with window size 400 and step size 40.
A – sample 44; B – sample 46; C – sample 13
Among the mutations associated with HIV resistance to non-nucleoside reverse transcriptase inhibitors, a relatively rare non-polymorphic V179T mutation has been shown, which is sometimes selected in patients receiving NNRTIs. This is due to the minimal decrease in the sensitivity of etravirine and rilpivirine [13, 14]. Several substitutions were also encountered in position 238: K238T, which reduces the susceptibility to nevirapine and efavirenz by about 5 times; K238R, which is a common polymorphism that does not reduce susceptibility to NNRTIs [15]; K238E is a rare mutation in this position, the effect of which is not well described in the literature.
Выявленные мутации, ассоциированные с лекарственной устойчивостью, как возникающие спонтанно, так и вследствие передачи от пациентов с вирусологической неэффективностью, могут активно распространяться среди новых пациентов, вследствие низкой информированности населения о ВИЧ-инфекции, труднодоступности медицинской помощи и средств контрацепции. Следствием низкой информированности о гемоконтактных инфекциях может быть повышен травматический риск передачи вируса, возможность которого показана в различных исследованиях (ссылка на нас и еще-кого-нибудь желательно)
В заключение необходимо отметить, что даже совместный анализ данных, полученных в ходе данного исследования и в работе 2016 года, недостаточен для того, чтобы в полной мере изучить генетическое разнообразие ВИЧ в Гвинейской республике. Необходимы более подробные исследования на больших выборках для того, чтобы сделать заключение о генетической структуре вируса на территории со столь сложной эпидемиологической ситуацией.
Table 2. Complete name of samples and reference sequences in phylogenetic tree (fig. 2)
|
Complete sample name
Subtype in REGA v.3.0
| Subtype in phylogenetic analysis
| | 1
| 4352
| A1
| A1
|
| 2
| 6397
| G
| G
|
| 3
| 1128
| A1
| A1
|
| 4
| 3879
| A1
| A3
|
| 5
| 6436
| A1
| A1
|
| 6
| 4238
| A1
| A1
|
| 7
| 3169
| A1
| CRF 02AG
|
| 8
| 1786
| CRF 02AG
| CRF 02AG
|
| 9
| 4886
| CRF 02AG
| CRF 02AG
|
| 10
| 2434
| CRF 02AG
| CRF 02AG
|
| 11
| 1912
| CRF 02AG
| CRF 02AG
|
| 12
| 5721
| Recombinant 02AG/A1
| CRF 02AG
|
| 13
| 1286
| Recombinant G/A1
| G
|
| 14
| 2492
| CRF 02AG
| CRF 02AG
|
| 15
| 1037
| HIV A
| A6
|
| 16
| 1048
| CRF 02AG
| CRF 02AG
|
| 17
| 2356
| CRF 02AG
| CRF 02AG
|
| 18
| 1170
| Recombinant G/A1
| Recombinant G/A1
|
| 19
| 1156
| CRF 02AG
| CRF 02AG
|
| 20
| 1260
| CRF 02AG
| CRF 02AG
|
| 21
| 2709
| Recombinant 02AG/A1
| CRF 02AG
|
| 22
| 1036
| Recombinant 02AG/A1
| CRF 02AG
|
| 23
| 2886
| CRF 02AG
| CRF 02AG
|
| 24
| 2595
| CRF 02AG
| CRF 02AG
|
| 25
| 2141
| CRF 02AG
| CRF 02AG
|
| 26
| 2046
| CRF 02AG
| CRF 02AG
|
| 27
| 1969
| A
| A1
|
| 28
| 1976
| A
| A3
|
| 29
| 2073
| G-like
| G
|
| 30
| 2813
| CRF 02AG
| CRF 02AG
|
| 31
| 2205
| CRF 02AG
| CRF 02AG
|
| 32
| 2501
| CRF 06CPX
| CRF 06CPX
|
| 33
| 2731
| CRF 06CPX
| CRF 06CPX
|
| 34
| 2279
| CRF 06CPX
| CRF 06CPX
|
| 35
| 2488
| CRF 02AG
| CRF 02AG
|
| 36
| 2130
| A1
| A3
|
| 37
| 2504
| CRF 02AG
| CRF 02AG
|
| 38
| 2505
| CRF 02AG
| CRF 02AG
|
| 39
| 2506
| CRF 02AG
| CRF 02AG
|
| 40
| 2526
| CRF 02AG
| CRF 02AG
|
| 41
| 2530
| A
| A1
|
| 42
| 2547
| A
| A3
|
| 43
| 2562
| CRF 02AG
| CRF 02AG
|
| 44
| 2564
| Recombinant 02AG/A1
| CRF 02AG
|
| 45
| 1261
| A1
| A3
|
| 46
| 2664
| Recombinant 02AG/A1
| CRF 02AG
|
| 47
| 1215
| A1
| A3
|
| 48
| 1333
| Recombinant 14BG/G
| G
|
| 49
| 3816
| A
| A3
|
| 50
| 2670
| G
| G
|
| 51
| 2412
| CRF 02AG
| CRF 02AG
|
| 52
| 2578
| CRF 02AG
| CRF 02AG
|
| 53
| 6778
| A
| A3
|
| 54
| 1762
| CRF 02AG
| CRF 02AG
|
| 55
| 7032
| CRF 02AG
| CRF 02AG
|
| 56
| 1805
| CRF 02AG
| CRF 02AG
|
| 57
| 2309
| CRF 02AG
| CRF 02AG
|
| 58
| 954
| A
| A1
|
| ref1_A3
| AB098332
| A3
| A3
|
| ref2_CRF02_AG
| AB231896
| CRF 02AG
| CRF 02AG
|
| ref3__CRF02_AG
| AB231898
| CRF 02AG
| CRF 02AG
|
| ref4_A1
| AB287376
| A1
| A1
|
| ref5_G
| AF061641
| G
| G
|
| ref6_CRF02_AG
| AF063224
| CRF 02AG
| CRF 02AG
|
| ref7_C
| AF067155
| C
| C
|
| ref8_A1
| AF069670
| A1
| A1
|
| ref9_F1
| AF075703
| F1
| F1
|
| ref10_G
| AF084936
| G
| G
|
| ref11_A
| AF107771
| A
| A
|
| ref12_CRF03_AB
| AF193276
| CRF 03AB
| CRF 03AB
|
| ref13_A2
| AF286237
| A2
| A2
|
| ref14_CRF02_AG
| AF377954
| CRF 02AG
| CRF 02AG
|
| ref15_A6
| AF413987
| A6
| A1
|
| ref16_A1
| AF484509
| A1
| A1
|
| ref17_CRF02_AG
| AY151001
| CRF 02AG
| CRF 02AG
|
| ref18_B
| AY173951
| B
| B
|
| ref19_A6
| AY500393
| A6
| A6
|
| ref20_A3
| AY521629
| A3
| A3
|
| ref21_A3
| AY521631
| A3
| A3
|
| ref22_B
| AY713409
| B
| B
|
| ref23_C
| AY772699
| C
| C
|
| ref24_A6
| EF589043
| A6
| A6
|
| ref25_A1
| EU110087
| A1
| A1
|
| ref26_CRF02_AG
| EU786671
| CRF 02AG
| CRF 02AG
|
| ref27_A1
| EU861977
| A1
| A1
|
| ref28__CRF02_AG
| GU201514
| CRF 02AG
| CRF 02AG
|
| ref29_B
| HM586190
| B
| B
|
| ref30_A6
| HQ161930
| A6
| A6
|
| ref31_A6
| HQ449397
| A6
| A6
|
| ref32_B
| KJ771697
| B
| B
|
| ref33_CRF02_AG
| KT124792
| CRF 02AG
| CRF 02AG
|
| ref34_B
| M17449
| B
| B
|
| ref35_C
| U46016
| C
| C
|
| ref36_A1
| U51190
| A1
| A1
|
| ref37_C
| U52953
| C
| C
|
| ref38_G
| U88826
| G
| G
|
| ref39_CRF_06cpx
| MH605500.1
| CRF 06cpx
| CRF 06cpx
|
| ref40_CRF_06cpx
| HQ529257.1
| CRF 06cpx
| CRF 06cpx
|
References
1. UNAIDS data 2020. UNAIDS. Jul 2020 https://www.unaids.org/en/resources/documents/2020/unaids-data
2. UNAIDS data 2020. UNAIDS. Jul 2020
https://www.unaids.org/en/resources/documents/2020/unaids-data
3. Bekolo CE, Soumah MM, Tiemtore OW, et al. Assessing the outcomes of HIV-infected persons receiving treatment for Kaposi sarcoma in Conakry-Guinea. BMC Cancer. 2017;17(1):806. Published 2017 Dec 2. doi:10.1186/s12885-017-3771-x
4. Bbosa N, Kaleebu P, Ssemwanga D. HIV subtype diversity worldwide. Curr Opin HIV AIDS. 2019;14(3):153-160. DOI:10.1097/COH.0000000000000534
5. Lihana RW, Ssemwanga D, Abimiku A, Ndembi N. Update on HIV-1 diversity in Africa: a decade in review. AIDS Rev 2012; 14:83–100.
6. Nii-Trebi, Nicholas & Brandful, James & Ibe, Shiro & Sugiura, Wataru & Barnor, Jacob & Bampoh, Patrick & Yamaoka, Shoji & Matano, Tetsuro & Yoshimura, Kazuhisa & Ishikawa, Koichi & Ampofo, William. (2017). Dynamic HIV-1 genetic recombination and genotypic drug resistance among treatment-experienced adults in northern Ghana. Journal of Medical Microbiology. 66. DOI: 10.1099/jmm.0.000621.
7. Mbange AE, Kaba D, Diouara AAM, Diop-Ndiaye H, Ngom-Ngueye NF, Dieng A, Lo S, Toure KN, Fall M, Mbacham WF, Diallo MS, Cisse M, Mboup S, Kane CT. Surveillance of transmitted HIV-1 antiretroviral drug resistance in the context of decentralized HIV care in Senegal and the Ebola outbreak in Guinea. BMC Res Notes. 2018 Oct 12;11(1):723. doi: 10.1186/s13104-018-3804-9. PMID: 30309385; PMCID: PMC6182815.
8. Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016; 33(7): 1870–4. DOI: 10.1093/molbev/msw054.
9. Svarovskaia, Evguenia & Feng, Joy & Margot, Nicolas & Myrick, Florence & Goodman, Derrick & ly, John & White, Kirsten & Kutty, Nilima & Wang, Ruth & Borroto-Esoda, Katyna & Miller, Michael. (2008). The A62V and S68G Mutations in HIV-1 Reverse Transcriptase Partially Restore the Replication Defect Associated With the K65R Mutation. Journal of acquired immune deficiency syndromes (1999). 48. 428-36. DOI:10.1097/QAI.0b013e31817bbe93.
10. Yendewa G.A., Sahr F., Lakoh S., Ruiz M., Patiño L., Tabernilla A., Deen G.F., Sesay M., Salata R.A., Poveda E. Prevalence of drug resistance mutations among ART-naive and -experienced HIV-infected patients in Sierra Leone. J. Antimicrob. Chemother. 2019; 74(7):2024–9. DOI: 10.1093/jac/dkz134.
11. Vingerhoets J., Peeters M., Azijn H., Tambuyzer L., Hoogstoel A., Nijs S., de Bethune M-P., Picchio G. An update of the list of NNRTI mutations associated with decreased virological response to etravirine: multivariate analyses on the pooled DUET-1 and DUET-2 clinical trial data [abstract 24]. Antiviral Therapy. 2008; 13 Suppl 3:A26.
12. Flor-Parra F, Pérez-Pulido AJ, Pachón J, Pérez-Romero P. The HIV type 1 protease L10I minor mutation decreases replication capacity and confers resistance to protease inhibitors. AIDS Res Hum Retroviruses. 2011;27(1):65-70. DOI:10.1089/aid.2010.0072
13. Vingerhoets J., Peeters M., Azijn H., Tambuyzer L., Hoogstoel A., Nijs S., de Bethune M-P., Picchio G. An update of the list of NNRTI mutations associated with decreased virological response to etravirine: multivariate analyses on the pooled DUET-1 and DUET-2 clinical trial data [abstract 24]. Antiviral Therapy. 2008; 13 Suppl 3:A26.
14. John P. Barnard, Kelly D. Huber, Nicolas Sluis-Cremer. Nonnucleoside Reverse Transcriptase Inhibitor Hypersusceptibility and Resistance by Mutation of Residue 181 in HIV-1 Reverse Transcriptase. Antimicrobial Agents and Chemotherapy Jul 2019, 63 (8); DOI:10.1128/AAC.00676-19
15. Tambuyzer, L., H. Azijn, L. T. Rimsky, J. Vingerhoets, P. Lecocq, G. Kraus, G. Picchio, and M. P. de Bethune. 2009. Compilation and prevalence of mutations associated with resistance to non-nucleoside reverse transcriptase inhibitors. Antivir. Ther.14:103-109
Abstract
Objective: analysis of primary drug resistance of HIV in adult patients from the Republic of Guinea
Design: Cross-sectional study.
Methods: 2,168 residents of the Republic of Guinea (blood donors, as well as Rusal employees and their families who were undergoing routine medical examination) were tested for the presence of HIV serological markers using ELISA. Individuals with a positive result were further examined for the presence of viral load (VL) in blood plasma.
If VL was high enough (> 500 copies / ml), HIV was tested using Sanger sequencing. The obtained sequences were genotyped using the REGA HIV-1 Subtyping Tool - Version 3.0 and phylogenetic analysis in the MEGA 10 program. Analysis for the presence of mutations associated with drug resistance was carried out using the Stanford University HIV Drug Resistance Database.
Results: Serological markers of HIV were detected in 239 people, which was 11.02% of the complete sample. HIV RNA was detected in 58 people.
Genotyping revealed the prevalence of circulating recombinant HIV CRF02_AG (41.9%) in the study group, as well as other subtypes: A1 (29.1%); A3 (12.9%); non-CRF02_AG recombinant between A1 and G (12.9%); subtype G (3.2%).
In 25% of patients, at least one significant mutation was encountered, leading directly to drug resistance of HIV for their virus subtype. The mutations encountered cause resistance to nucleoside and non-nucleoside reverse transcriptase inhibitors; one case of multiple resistance was identified. Major resistance to IP was not met.
Conclusions: Thus, the genetic structure of HIV in the Republic of Guinea is close to that described in the literature for West African countries. The incidence of mutations associated with HIV resistance to ARVs was relatively high. Given the increasing number of patients who start taking ART, the high incidence of primary drug resistance can lead to frequent cases of therapy failure and, as a result, change of treatment regimens. To prescribe effective treatment regimens, it is necessary to introduce studies on the presence of primary resistance in newly diagnosed patients.
Key words: HIV-1 transmitted drug resistance, West Africa, Guinea Republic
Introduction
The human immunodeficiency virus (HIV) epidemic continues to spread rapidly around the world. According to The Joint United Nations Programme on HIV/AIDS (UNAIDS) estimates, currently the number of people infected with HIV on the Earth is about 31.6 - 44.5 million people, while the number of new infections amounted to 1.2 - 2.2 million cases in 2019 [1]. The African continent is one of the most HIV-affected territories in the world, currently home to 25.6 million people living with HIV (PLHIV), which is 67.37% of all registered HIV-infected in the world. The overwhelming majority (80.86%) are in the countries of Eastern and Southern Africa [2].
The Republic of Guinea is a located in West Africa. The HIV prevalence in Guinea was approximately 1.6% in 2014 and has practically unchanged by 2019, but treatment coverage also remains one of the lowest in the world, with less than a quarter (23%) of people living with HIV on antiretroviral therapy (ART) [1, 3].
Among other things, Africa, especially Western and Central, has the highest diversity of HIV subtypes in the world. In West and Central Africa, the most common circulating recombinant form (CRF) 02_AG. The A and G subtypes co-circulate in some countries such as Nigeria [4, 5, 6]. The study in 2016 confirmed the prevalence of CRF 02_AG in Guinea [7].
The Republic of Guinea is one of the countries affected by the Ebola epidemic, and therefore in the period 2014-2015, there were significant difficulties in providing treatment to patients with HIV infection. This, in turn, could affect the prevalence of drug resistance (DR), as a result of which more careful monitoring of HIV resistance is required in this region. However, the most recent data on drug resistance in Guinea reported in the literature dates back to 2009, when the prevalence of primary drug resistance was 8.9 % [ссылка]. In 2016, it was not possible to analyze a sufficient number of samples to identify the prevalence level of DR in the Republic of Guinea [7].
Methods
Study setting
During the study, 2,168 blood plasma samples were obtained from healthy people from the Republic of Guinea: blood donors and employees of UC RUSAL and their families with no data about disease caused by the Ebola virus. The surveyed persons denied a history of HIV infection. The study was conducted based of the Center of Epidemiology and Microbiology (CREMS) in Kindia. The material was collected by UC RUSAL employees and Institute of Applied Biological Research of Guinea (IRBAG) specialists.
Participant Enrollment
Inclusion criteria were: (i) ≥18 years (ii) no history of HIV infection
Laboratory Methods
Patients were examined for the presence of HIV antigens and antibodies to the virus by ELISA. Quantitative analysis of HIV RNA was carried out with a commercial kit "AmpliSens® HIV-Monitor-FRT" (Central Research Institute of Epidemiology, Russia) with a sensitivity threshold of 500 copies/ml, and samples with detectable viral load (VL) were analyzed using RT-PCR and Sanger sequencing.
For reverse transcription and amplification of HIV, commercial kits "RT-PCR-kit-Pro/Rev" and "PCR-kit-Pro/Rev" (Central Research Institute of Epidemiology, Russia) were used, the sequencing reaction was performed according to the instructions for the kit "AmpliSens® HIVResist-Seq" (Central Research Institute of Epidemiology, Russia) [как было показано ранее или типа того и ссылка на что-то из наших статей].
For HIV genotyping we used nucleotide sequences of 1302 nt spanning the pol gene in the range of 2253–3554 nt., Coordinates are given for the data presented in the international GenBank database HIV HXB2 (K03455.1). The analysis of the sequencing reaction products was performed using an ABI Prism 3500 genetic analyzer (Applied Biosystems, USA).
Nucleotide sequences were analyzed using the NCBI Blast program in comparison with nucleotide sequences presented in the international GenBank database. Alignment of nucleotide sequences was executed with MEGAv.7.0 program using the ClustalW algorithm [8]. For the construction of phylogenetic trees and subsequent phylogenetic analysis, the Neighbor-joining algorithm was used, which allows optimization of trees by the criterion of “balanced minimum evolution”; when assessing the reliability of phylogenetic relationships, we used multiple generations of samples using the Bootstrap method for 1000 independent constructions of each phylogenetic tree.
Isolates were genotyped using the REGA HIV-1 Subtyping Tool 3.0 program and analysis of their phylogenetic relationships with reference sequences from the GenBank international database (Fig. 1), which made it possible to more accurately assess the distribution of HIV-1 subtypes in the studied population. Analysis of HIV genetic sequences for the presence of drug resistance mutations was performed using the Stanford HIV Data Base
Results
Study Population
Суммарно был исследован материл от 2168 пациентов, возраст которых находился в пределах 18-58 лет, медианный возраст составил 38 лет (β = 0.95, (37.58; 39.72)). Большинство обследованных пациентов – мужского пола (68.17% (β = 0.95; (66.21%; 70.13%))).
Genotyped samples were obtained from patients aged 18 to 54 years. The median age was 36 years (β = 0.95, (32.53; 39.47)). The predominant age group is from 31 to 45 years old (62.07%). The most represented in the sample are men, they make up 63.79% of the study group (β = 0.95, (52%; 76%)) compared with women - 36.21% (β = 0.95, (24%; 49%)).

Fig 1. Demographic characteristics of antiretroviral-naive patients. M – male; F – female.